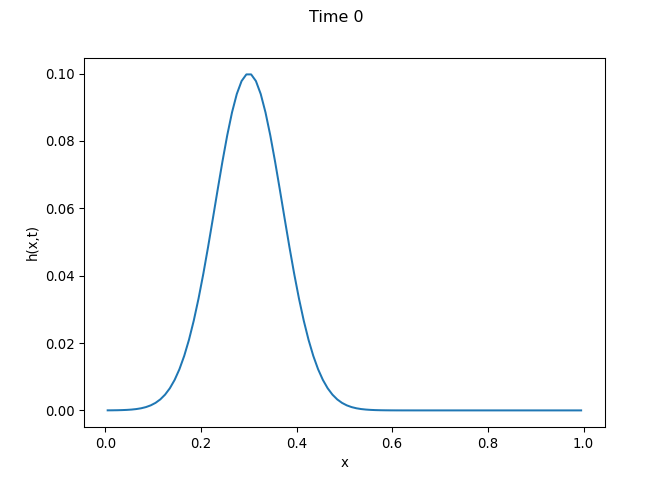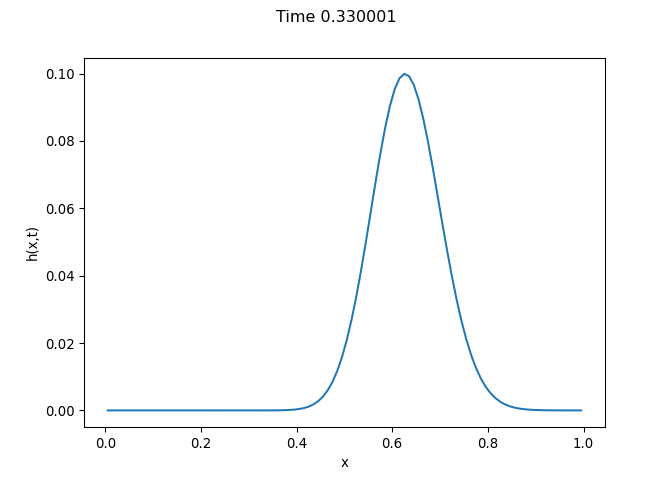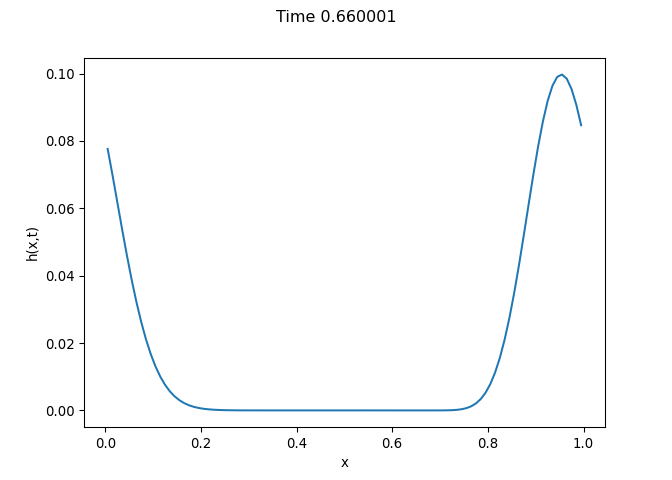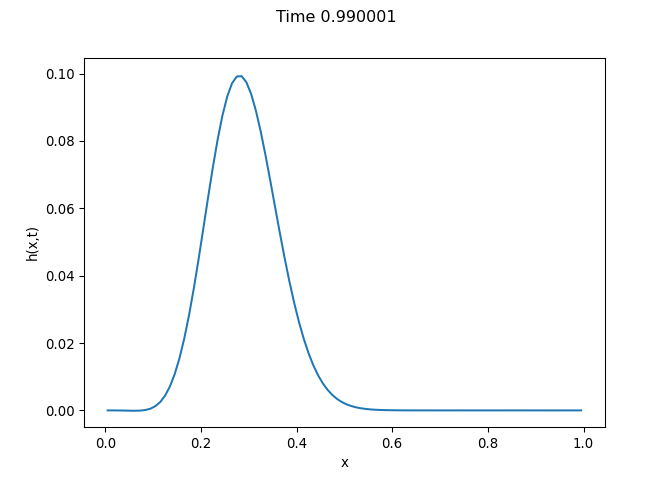
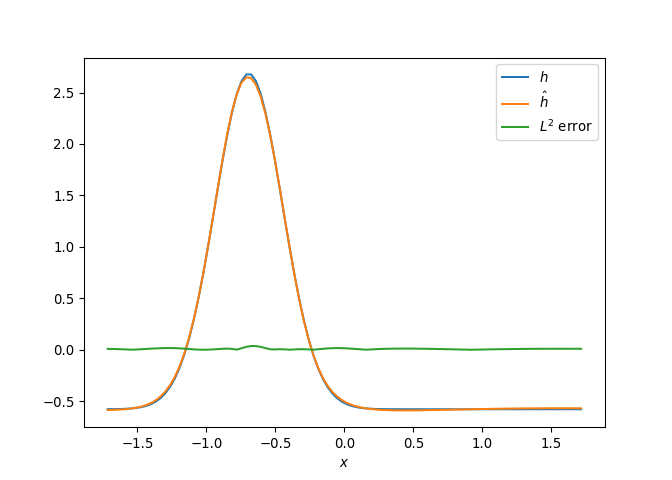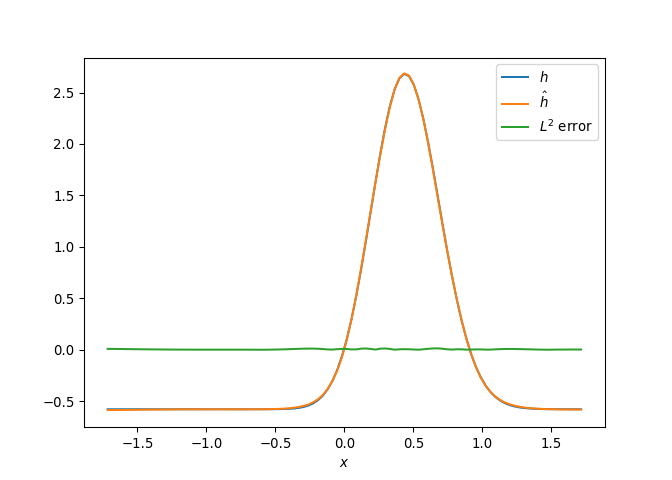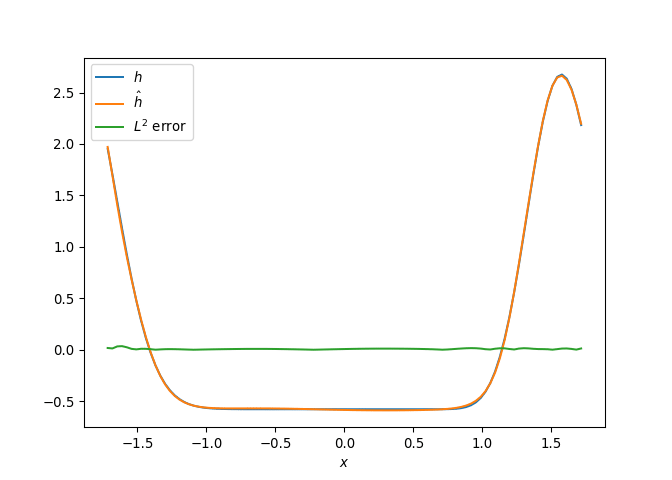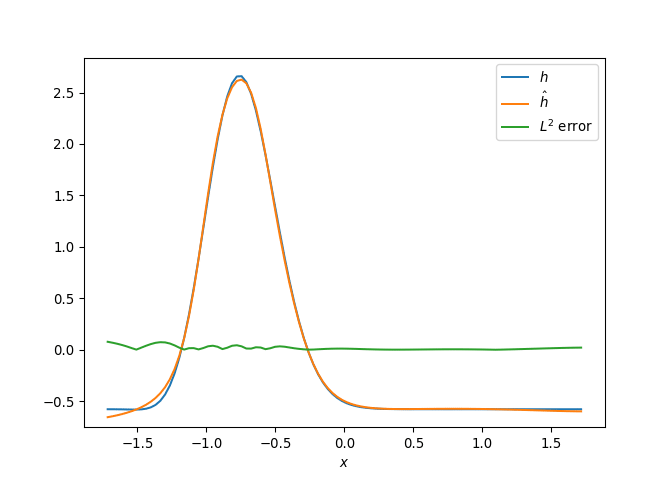
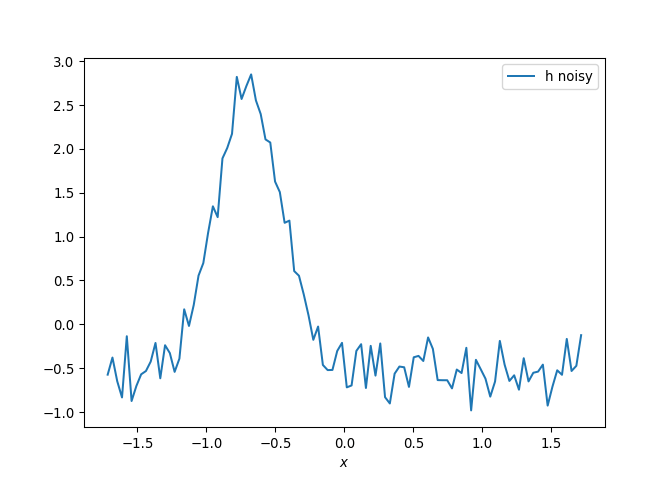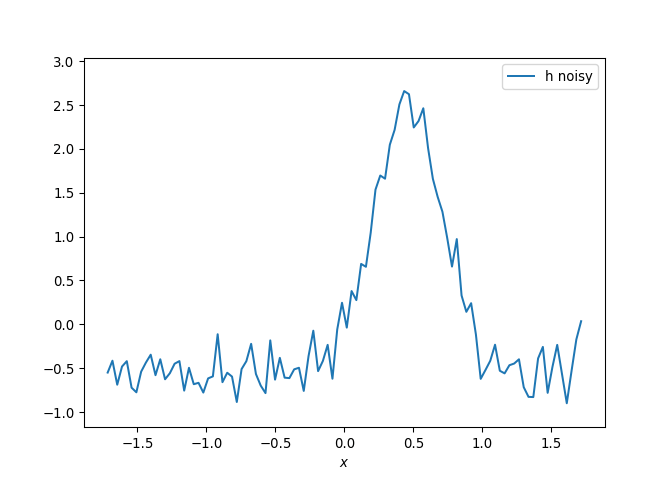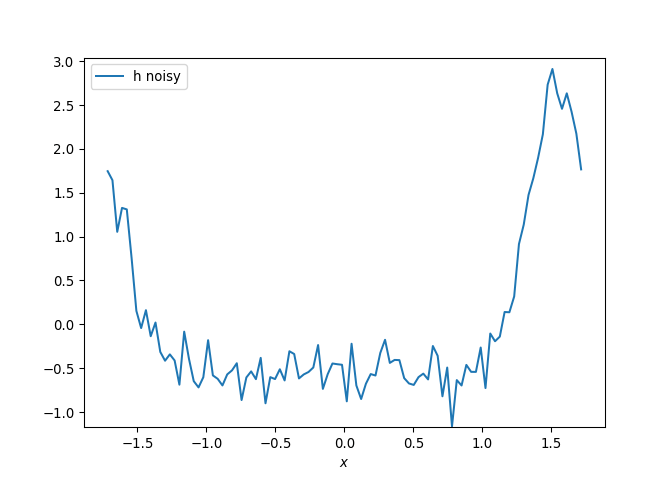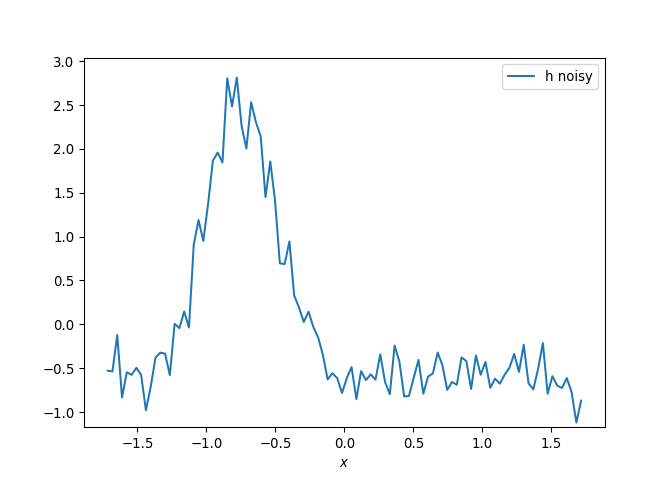
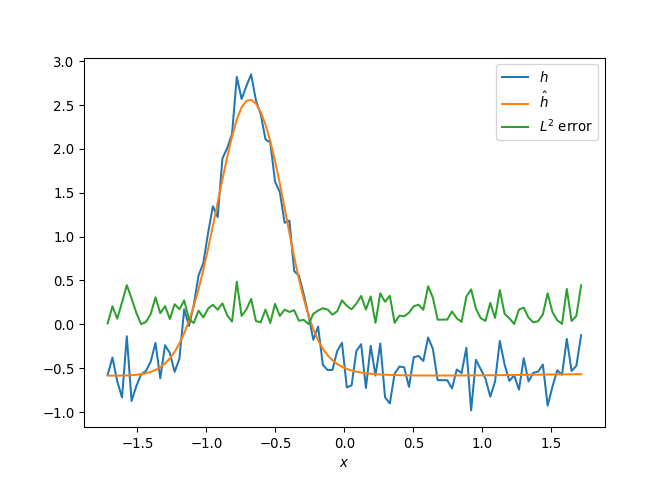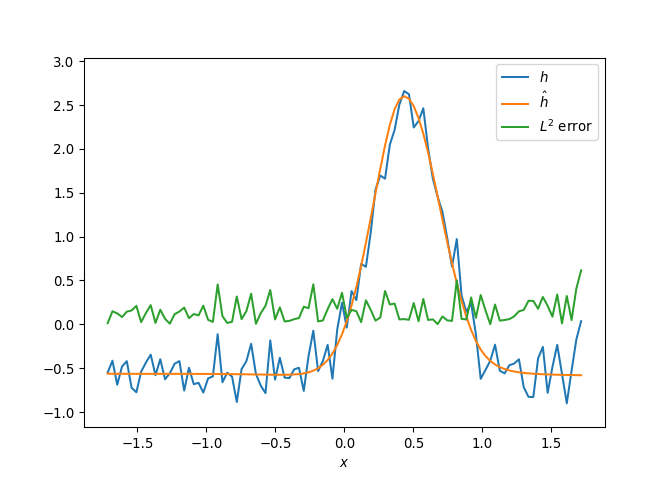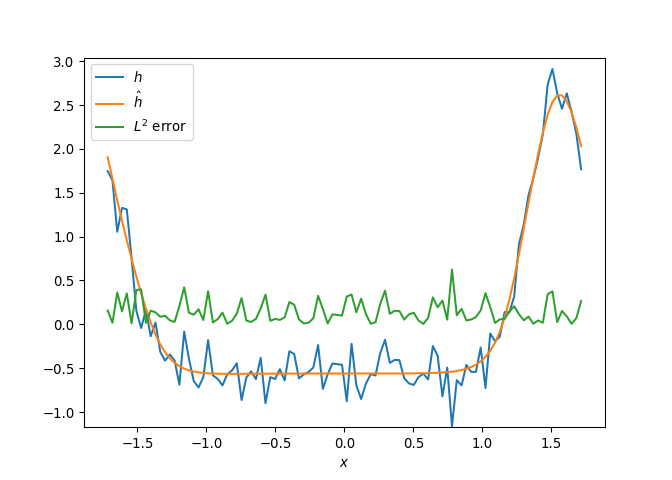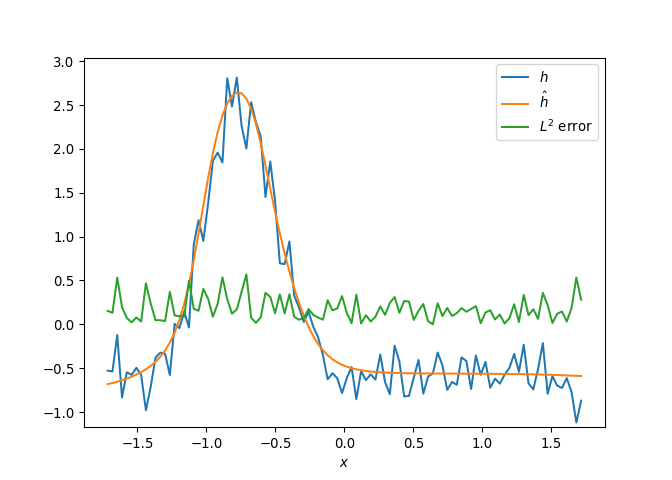
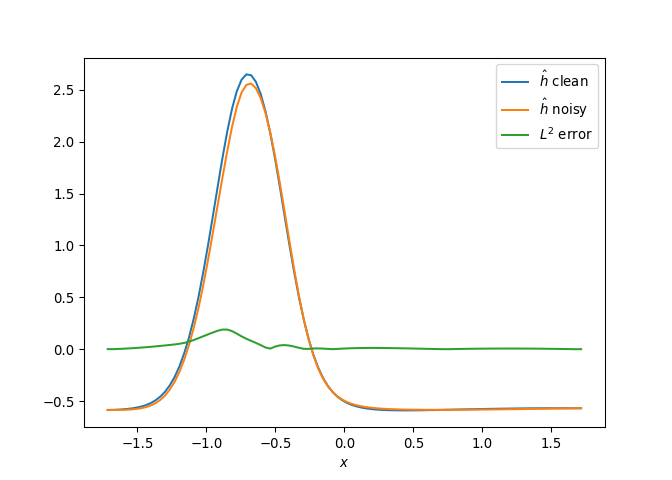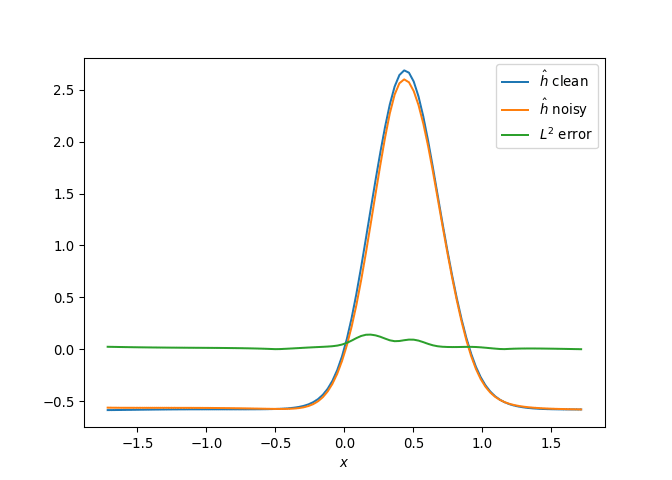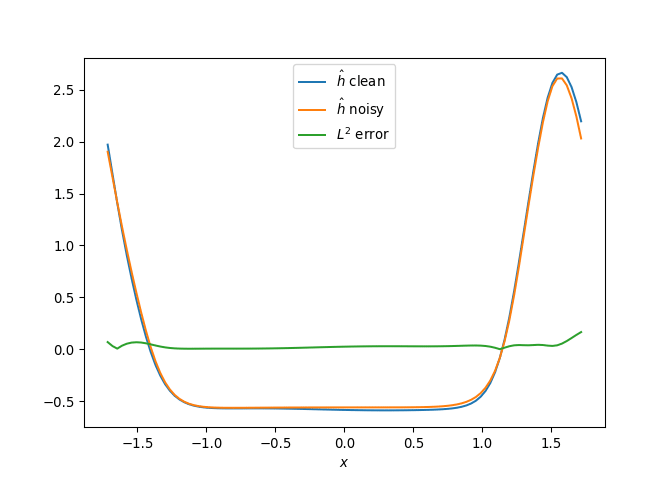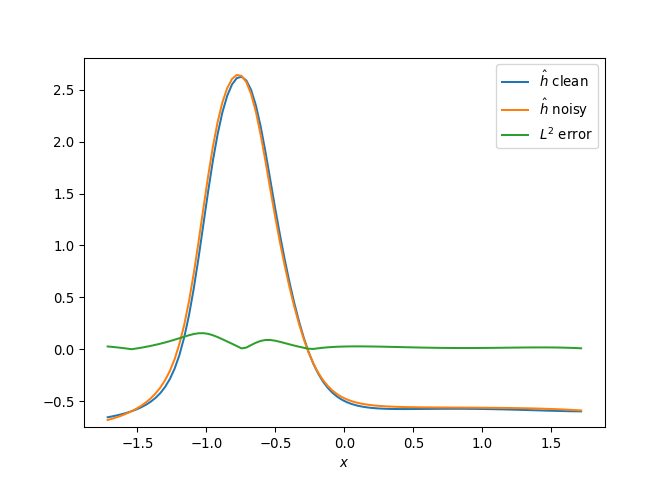
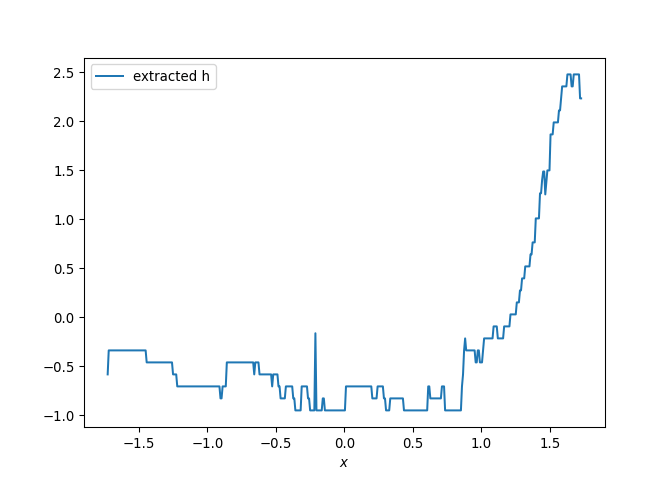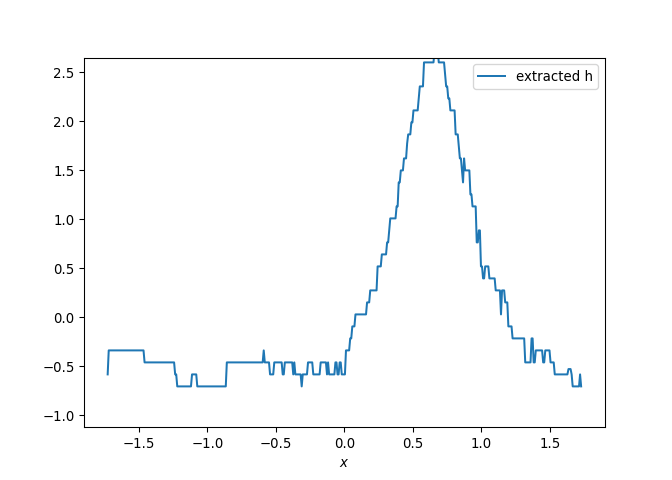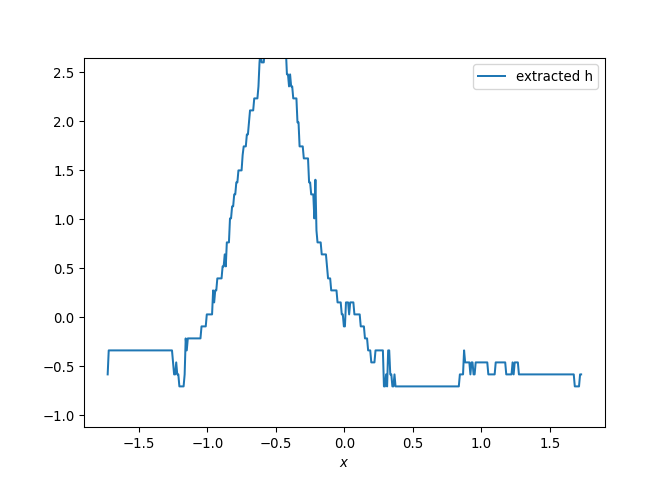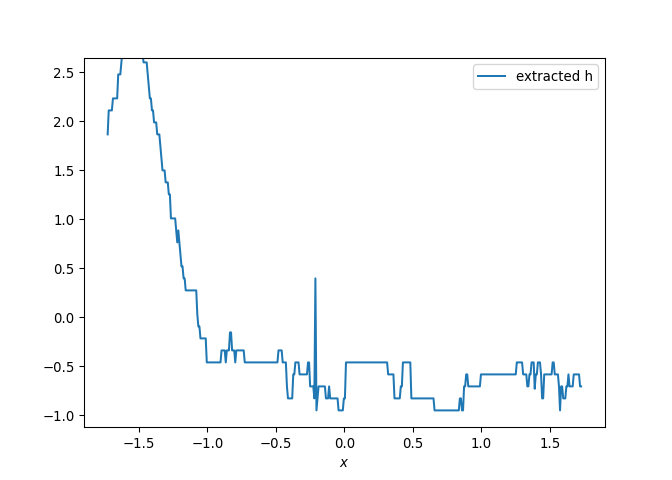
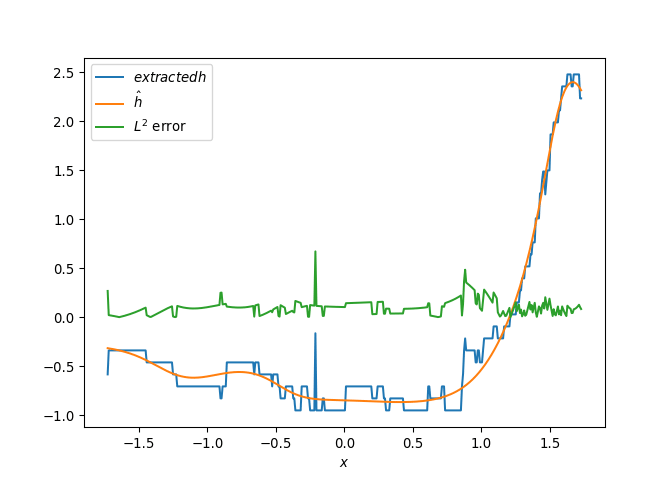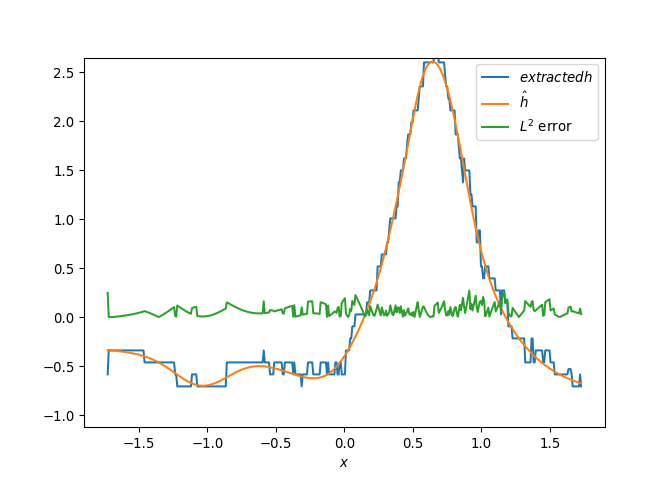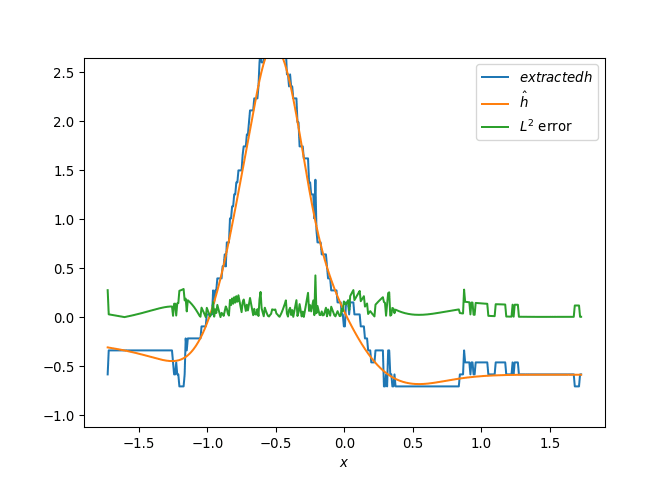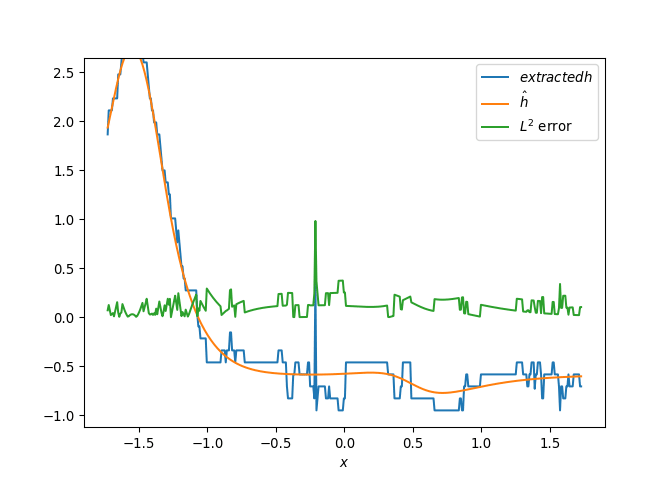
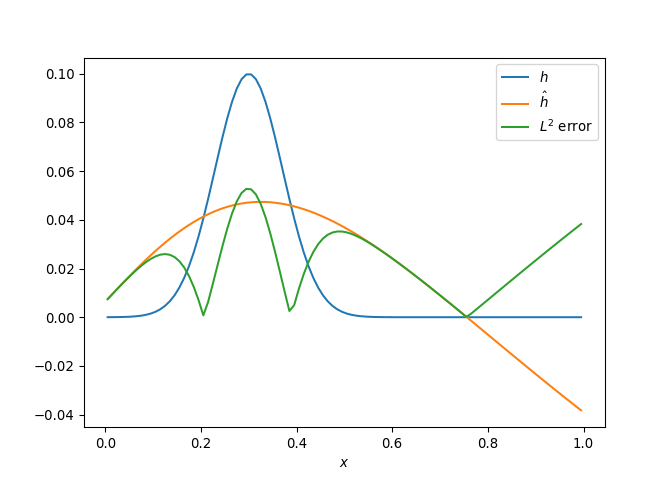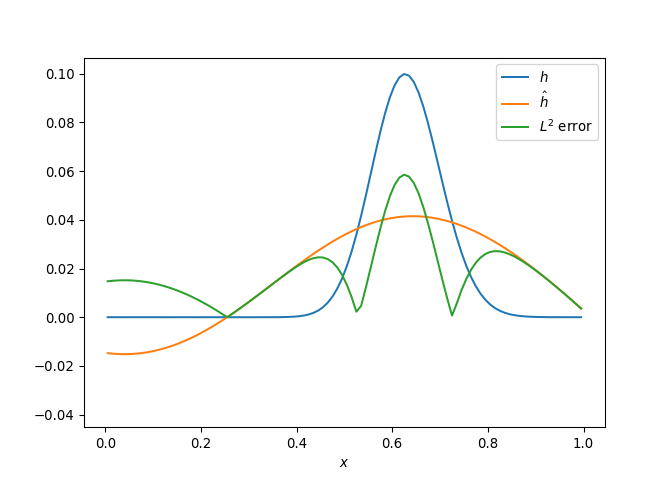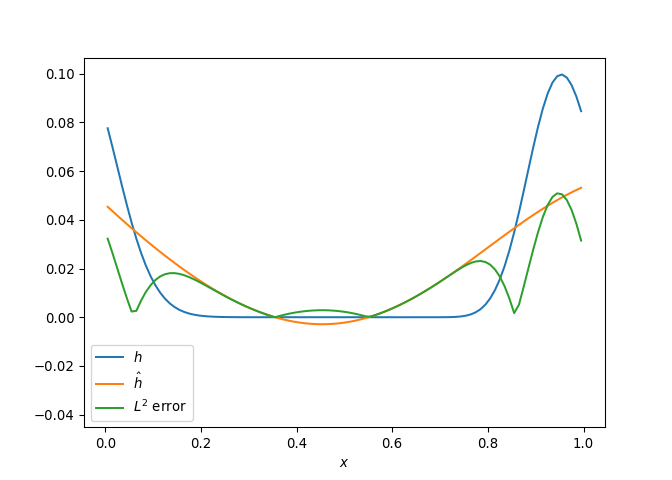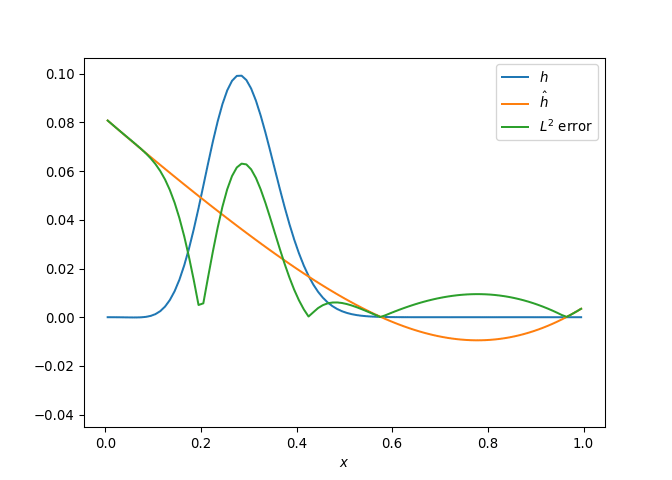
[h(x, t), h_x(x, t), h_xx(x, t), h_xxx(x, t), h_xxxx(x, t), h(x, t)**2, h(x, t)*h_x(x, t), h(x, t)*h_xx(x, t), h(x, t)*h_xxx(x, t), h(x, t)*h_xxxx(x, t), h_x(x, t)**2, h_x(x, t)*h_xx(x, t), h_x(x, t)*h_xxx(x, t), h_x(x, t)*h_xxxx(x, t), h_xx(x, t)**2, h_xx(x, t)*h_xxx(x, t), h_xx(x, t)*h_xxxx(x, t), h_xxx(x, t)**2, h_xxx(x, t)*h_xxxx(x, t), h_xxxx(x, t)**2, h(x, t)**3, h(x, t)**2*h_x(x, t), h(x, t)**2*h_xx(x, t), h(x, t)**2*h_xxx(x, t), h(x, t)**2*h_xxxx(x, t), h(x, t)*h_x(x, t)**2, h(x, t)*h_x(x, t)*h_xx(x, t), h(x, t)*h_x(x, t)*h_xxx(x, t), h(x, t)*h_x(x, t)*h_xxxx(x, t), h(x, t)*h_xx(x, t)**2, h(x, t)*h_xx(x, t)*h_xxx(x, t), h(x, t)*h_xx(x, t)*h_xxxx(x, t), h(x, t)*h_xxx(x, t)**2, h(x, t)*h_xxx(x, t)*h_xxxx(x, t), h(x, t)*h_xxxx(x, t)**2, h_x(x, t)**3, h_x(x, t)**2*h_xx(x, t), h_x(x, t)**2*h_xxx(x, t), h_x(x, t)**2*h_xxxx(x, t), h_x(x, t)*h_xx(x, t)**2, h_x(x, t)*h_xx(x, t)*h_xxx(x, t), h_x(x, t)*h_xx(x, t)*h_xxxx(x, t), h_x(x, t)*h_xxx(x, t)**2, h_x(x, t)*h_xxx(x, t)*h_xxxx(x, t), h_x(x, t)*h_xxxx(x, t)**2, h_xx(x, t)**3, h_xx(x, t)**2*h_xxx(x, t), h_xx(x, t)**2*h_xxxx(x, t), h_xx(x, t)*h_xxx(x, t)**2, h_xx(x, t)*h_xxx(x, t)*h_xxxx(x, t), h_xx(x, t)*h_xxxx(x, t)**2, h_xxx(x, t)**3, h_xxx(x, t)**2*h_xxxx(x, t), h_xxx(x, t)*h_xxxx(x, t)**2, h_xxxx(x, t)**3, h(x, t)**4, h(x, t)**3*h_x(x, t), h(x, t)**3*h_xx(x, t), h(x, t)**3*h_xxx(x, t), h(x, t)**3*h_xxxx(x, t), h(x, t)**2*h_x(x, t)**2, h(x, t)**2*h_x(x, t)*h_xx(x, t), h(x, t)**2*h_x(x, t)*h_xxx(x, t), h(x, t)**2*h_x(x, t)*h_xxxx(x, t), h(x, t)**2*h_xx(x, t)**2, h(x, t)**2*h_xx(x, t)*h_xxx(x, t), h(x, t)**2*h_xx(x, t)*h_xxxx(x, t), h(x, t)**2*h_xxx(x, t)**2, h(x, t)**2*h_xxx(x, t)*h_xxxx(x, t), h(x, t)**2*h_xxxx(x, t)**2, h(x, t)*h_x(x, t)**3, h(x, t)*h_x(x, t)**2*h_xx(x, t), h(x, t)*h_x(x, t)**2*h_xxx(x, t), h(x, t)*h_x(x, t)**2*h_xxxx(x, t), h(x, t)*h_x(x, t)*h_xx(x, t)**2, h(x, t)*h_x(x, t)*h_xx(x, t)*h_xxx(x, t), h(x, t)*h_x(x, t)*h_xx(x, t)*h_xxxx(x, t), h(x, t)*h_x(x, t)*h_xxx(x, t)**2, h(x, t)*h_x(x, t)*h_xxx(x, t)*h_xxxx(x, t), h(x, t)*h_x(x, t)*h_xxxx(x, t)**2, h(x, t)*h_xx(x, t)**3, h(x, t)*h_xx(x, t)**2*h_xxx(x, t), h(x, t)*h_xx(x, t)**2*h_xxxx(x, t), h(x, t)*h_xx(x, t)*h_xxx(x, t)**2, h(x, t)*h_xx(x, t)*h_xxx(x, t)*h_xxxx(x, t), h(x, t)*h_xx(x, t)*h_xxxx(x, t)**2, h(x, t)*h_xxx(x, t)**3, h(x, t)*h_xxx(x, t)**2*h_xxxx(x, t), h(x, t)*h_xxx(x, t)*h_xxxx(x, t)**2, h(x, t)*h_xxxx(x, t)**3, h_x(x, t)**4, h_x(x, t)**3*h_xx(x, t), h_x(x, t)**3*h_xxx(x, t), h_x(x, t)**3*h_xxxx(x, t), h_x(x, t)**2*h_xx(x, t)**2, h_x(x, t)**2*h_xx(x, t)*h_xxx(x, t), h_x(x, t)**2*h_xx(x, t)*h_xxxx(x, t), h_x(x, t)**2*h_xxx(x, t)**2, h_x(x, t)**2*h_xxx(x, t)*h_xxxx(x, t), h_x(x, t)**2*h_xxxx(x, t)**2, h_x(x, t)*h_xx(x, t)**3, h_x(x, t)*h_xx(x, t)**2*h_xxx(x, t), h_x(x, t)*h_xx(x, t)**2*h_xxxx(x, t), h_x(x, t)*h_xx(x, t)*h_xxx(x, t)**2, h_x(x, t)*h_xx(x, t)*h_xxx(x, t)*h_xxxx(x, t), h_x(x, t)*h_xx(x, t)*h_xxxx(x, t)**2, h_x(x, t)*h_xxx(x, t)**3, h_x(x, t)*h_xxx(x, t)**2*h_xxxx(x, t), h_x(x, t)*h_xxx(x, t)*h_xxxx(x, t)**2, h_x(x, t)*h_xxxx(x, t)**3, h_xx(x, t)**4, h_xx(x, t)**3*h_xxx(x, t), h_xx(x, t)**3*h_xxxx(x, t), h_xx(x, t)**2*h_xxx(x, t)**2, h_xx(x, t)**2*h_xxx(x, t)*h_xxxx(x, t), h_xx(x, t)**2*h_xxxx(x, t)**2, h_xx(x, t)*h_xxx(x, t)**3, h_xx(x, t)*h_xxx(x, t)**2*h_xxxx(x, t), h_xx(x, t)*h_xxx(x, t)*h_xxxx(x, t)**2, h_xx(x, t)*h_xxxx(x, t)**3, h_xxx(x, t)**4, h_xxx(x, t)**3*h_xxxx(x, t), h_xxx(x, t)**2*h_xxxx(x, t)**2, h_xxx(x, t)*h_xxxx(x, t)**3, h_xxxx(x, t)**4]