Predicting bacterial growth in experimental images via recurrent neural networks
Over the last several decades, advances in genetic sequencing and identification have massively expanded the catalog of bacterial species and strains. This in turn has exposed the myriad of ways that communities of bacteria contribute to the health and proper function of the natural world. However, these communities are often made up of a variety of species and strains whose interactions are incredibly complex. Researchers have thus looked for new experimental and analytical tools to capture and model this complexity or to forecast its outcome. Recently, deep learning methods developed for fields as diverse as weather prediction or self-driving cars have presented a new angle with which to approach this analysis.
This project focused on predicting the growth of a heterogeneous culture of bacterial strains via experimental images of their interactions and a spatiotemporal convolutional recurrent neural network called PredRNN [1]. Specifically, the network was trained on sequences of images from microwell arrays which show the growth of two mutant strains of Pseudomonas aeruginosa in a single well (one which can kill the other) [2]. A sample of one of these sequences can be seen below. The network then outputs a predicted sequence of subsequent images, which we compared with the true images via popular image metrics such as Learned Perceptual Image Patch Similarity (LPIPS) as well as with biological metrics such as the number and size of colonies of each strain. The results of this work can be found in this article [3].
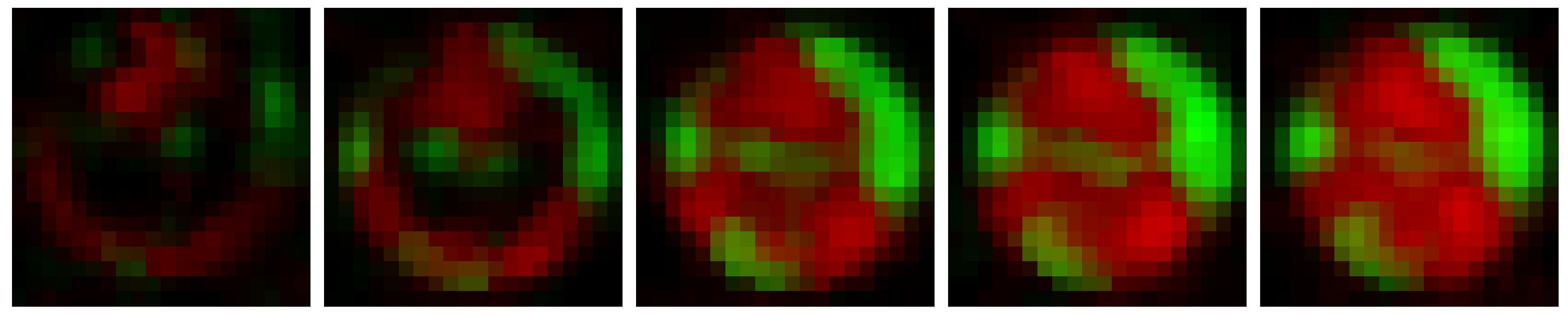
This same technique was also applied to accelerate the results of an agent based model of the same system which can be found in this preprint [4].